Introduction
The UCGD-Pipeline is our fully automated processing pipeline which at a high-level does the following:
- Uses DeepVariant based software following GATK best practice approach.
- Accepts input files of BAM/CRAM/Fastq or a combination of each.
- Utilizes multiple modern QC tools.
- Can use any number of background files for joint-genotyping.
- Processes using Nextflow.
- Uses container (Docker & Singularity) based software for HPC UCGD PE Processing.
- Uses and requires granted access via the U of U IRB system.
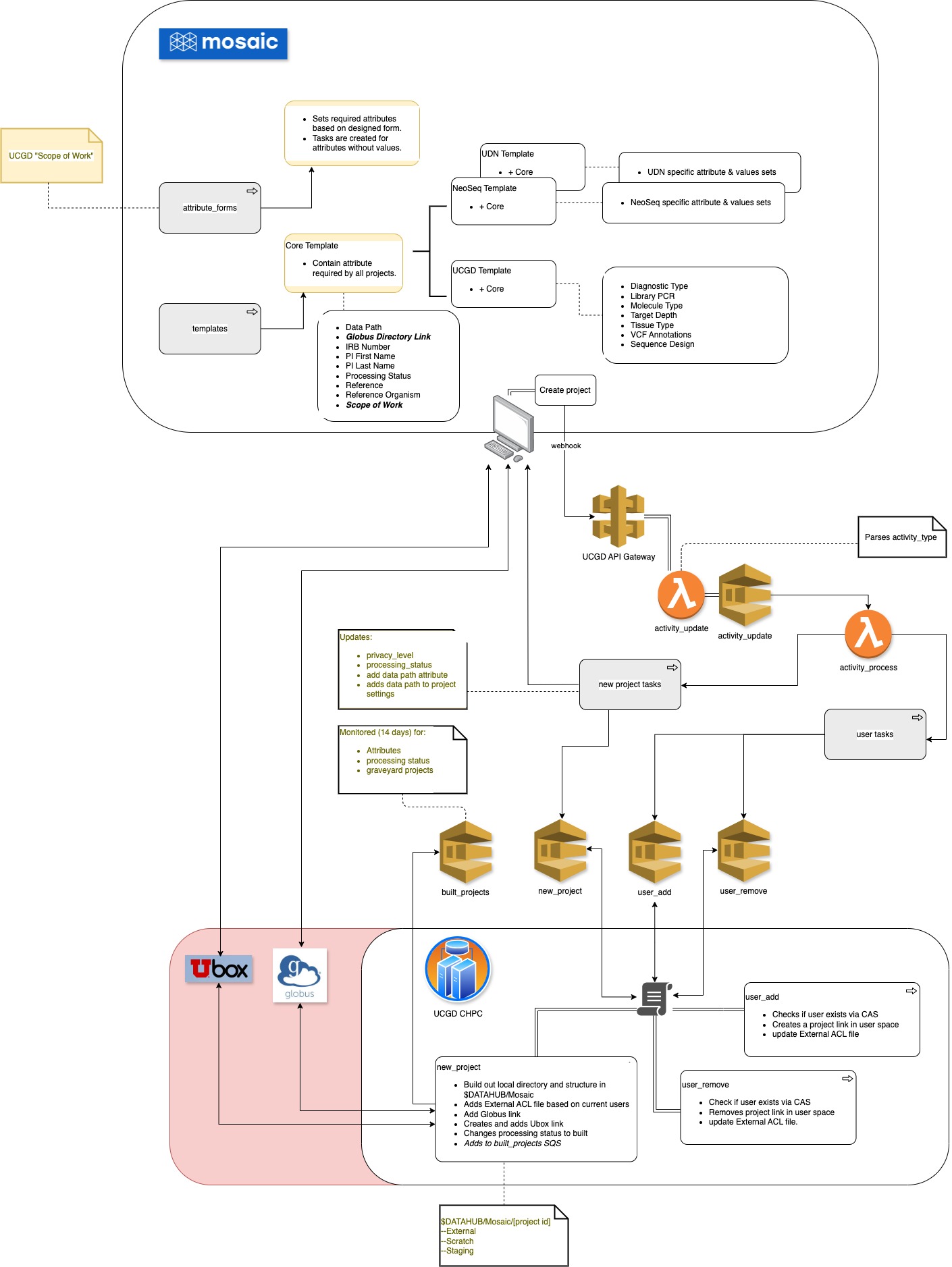
- Is automated using Mosaic for project LIMS management to determine when/how processing should be done.
- Manages and stored all project data within the CHPC Protected Environment.
Pipeline Overview
Adding New Software
Docker:
UCGD exclusively uses docker to manage all software for production level processing.
In order to add specific tools to the UCGD-Pipeline, we require a publicly accessible docker image (Nextflow cannot access private docker repositories at this time), that launches into a bash environment (see below).
Steps to add software or desired processes:
Adding tools to the pipeline is as easy as creating a docker container with the desired software available as an executable.
- Create a Docker container with the desired script/software executable via the container command line. i.e. no ENTRYPOINT.
- The container must also have the following commands required by nextflow awk, date, grep, egrep, ps, sed, tail, tee.
- Supply the desired command's to be run (either by example or SOP) by the containerized software which will be executed by nextflow.
- Detail list of final files which will be expected to be ready and accessible at process completion.
After we have been supplied with the above listed, the steps will be added as a automated processes.
Container Service:
UCGD uses Dockerhub to store all of our current containers, if you have an issue with publicly accessible containers please contact us.